heirarchical clustering

“Which of my samples are most closely related?”
So far we have been looking at how to plot raw data, summarize data, and reduce a data set’s dimensionality. It’s time to look at how to identify relationships between the samples in our data sets. For example: in the Alaska lakes dataset, which lake is most similar, chemically speaking, to Lake Narvakrak? Answering this requires calculating numeric distances between samples based on their chemical properties. For this, the first thing we need is a distance matrix:
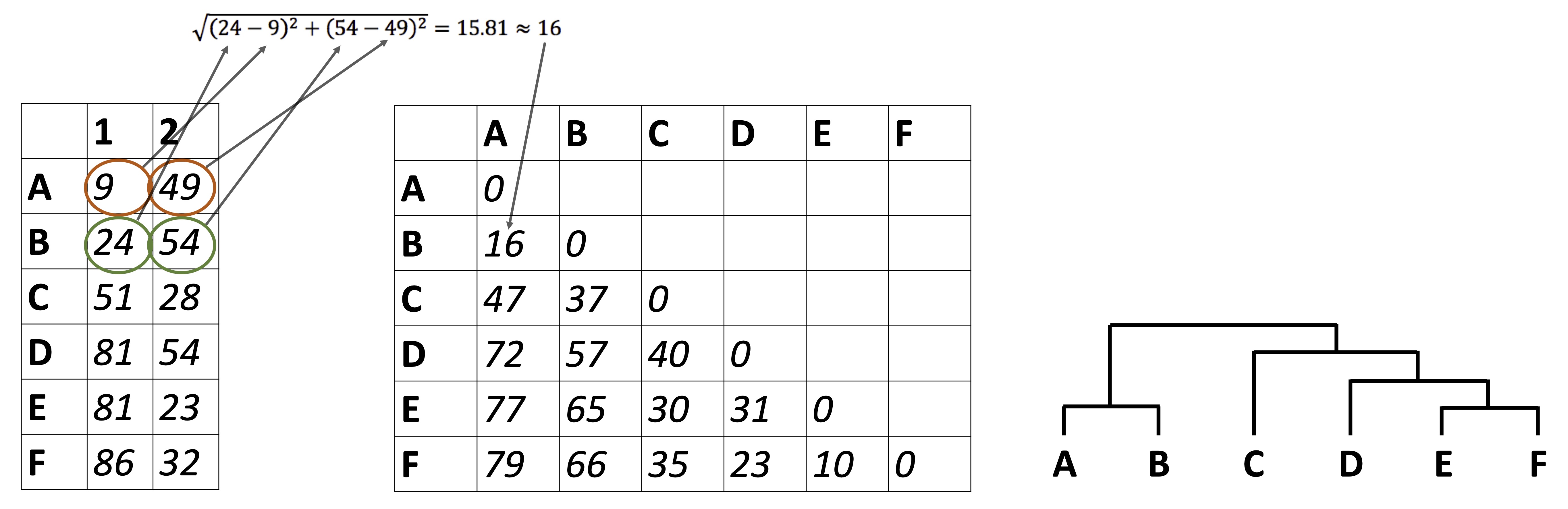
Please note that we can get distance matrices directly from runMatrixAnalysis by specifying analysis = "dist":
dist <- runMatrixAnalysis(
data = alaska_lake_data,
analysis = c("dist"),
column_w_names_of_multiple_analytes = "element",
column_w_values_for_multiple_analytes = "mg_per_L",
columns_w_values_for_single_analyte = c("water_temp", "pH"),
columns_w_additional_analyte_info = "element_type",
columns_w_sample_ID_info = c("lake", "park")
)
## Replacing NAs in your data with mean
as.matrix(dist)[1:3,1:3]
## Devil_Mountain_Lake_BELA
## Devil_Mountain_Lake_BELA 0.000000
## Imuruk_Lake_BELA 3.672034
## Kuzitrin_Lake_BELA 1.663147
## Imuruk_Lake_BELA
## Devil_Mountain_Lake_BELA 3.672034
## Imuruk_Lake_BELA 0.000000
## Kuzitrin_Lake_BELA 3.062381
## Kuzitrin_Lake_BELA
## Devil_Mountain_Lake_BELA 1.663147
## Imuruk_Lake_BELA 3.062381
## Kuzitrin_Lake_BELA 0.000000There is more that we can do with distance matrices though, lots more. Let’s start by looking at an example of hierarchical clustering. For this, we just need to tell runMatrixAnalysis() to use analysis = "hclust":
AK_lakes_clustered <- runMatrixAnalysis(
data = alaska_lake_data,
analysis = "hclust",
column_w_names_of_multiple_analytes = "element",
column_w_values_for_multiple_analytes = "mg_per_L",
columns_w_values_for_single_analyte = c("water_temp", "pH"),
columns_w_additional_analyte_info = "element_type",
columns_w_sample_ID_info = c("lake", "park"),
na_replacement = "mean"
)
AK_lakes_clustered
## # A tibble: 39 × 26
## sample_unique_ID lake park parent node branch.length
## <chr> <chr> <chr> <int> <int> <dbl>
## 1 Devil_Mountain_La… Devi… BELA 33 1 8.12
## 2 Imuruk_Lake_BELA Imur… BELA 32 2 4.81
## 3 Kuzitrin_Lake_BELA Kuzi… BELA 37 3 3.01
## 4 Lava_Lake_BELA Lava… BELA 38 4 2.97
## 5 North_Killeak_Lak… Nort… BELA 21 5 254.
## 6 White_Fish_Lake_B… Whit… BELA 22 6 80.9
## 7 Iniakuk_Lake_GAAR Inia… GAAR 29 7 3.60
## 8 Kurupa_Lake_GAAR Kuru… GAAR 31 8 8.57
## 9 Lake_Matcharak_GA… Lake… GAAR 29 9 3.60
## 10 Lake_Selby_GAAR Lake… GAAR 30 10 4.80
## # ℹ 29 more rows
## # ℹ 20 more variables: label <chr>, isTip <lgl>, x <dbl>,
## # y <dbl>, branch <dbl>, angle <dbl>, bootstrap <dbl>,
## # water_temp <dbl>, pH <dbl>, C <dbl>, N <dbl>, P <dbl>,
## # Cl <dbl>, S <dbl>, F <dbl>, Br <dbl>, Na <dbl>,
## # K <dbl>, Ca <dbl>, Mg <dbl>It works! Now we can plot our cluster diagram with a ggplot add-on called ggtree. We’ve seen that ggplot takes a “data” argument (i.e. ggplot(data = <some_data>) + geom_*() etc.). In contrast, ggtree takes an argument called tr, though if you’re using the runMatrixAnalysis() function, you can treat these two (data and tr) the same, so, use: ggtree(tr = <output_from_runMatrixAnalysis>) + geom_*() etc.
Note that ggtree also comes with several great new geoms: geom_tiplab() and geom_tippoint(). Let’s try those out:
library(ggtree)
AK_lakes_clustered %>%
ggtree() +
geom_tiplab() +
geom_tippoint() +
theme_classic()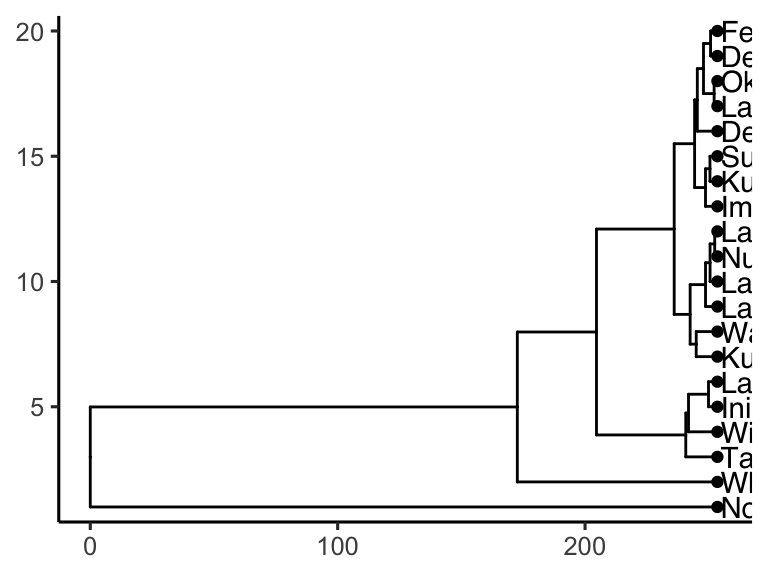
Cool! Though that plot could use some tweaking… let’s try:
AK_lakes_clustered %>%
ggtree() +
geom_tiplab(aes(label = lake), offset = 10, align = TRUE) +
geom_tippoint(shape = 21, aes(fill = park), size = 4) +
scale_x_continuous(limits = c(0,375)) +
scale_fill_brewer(palette = "Set1") +
# theme_classic() +
theme(
legend.position = c(0.2,0.8)
)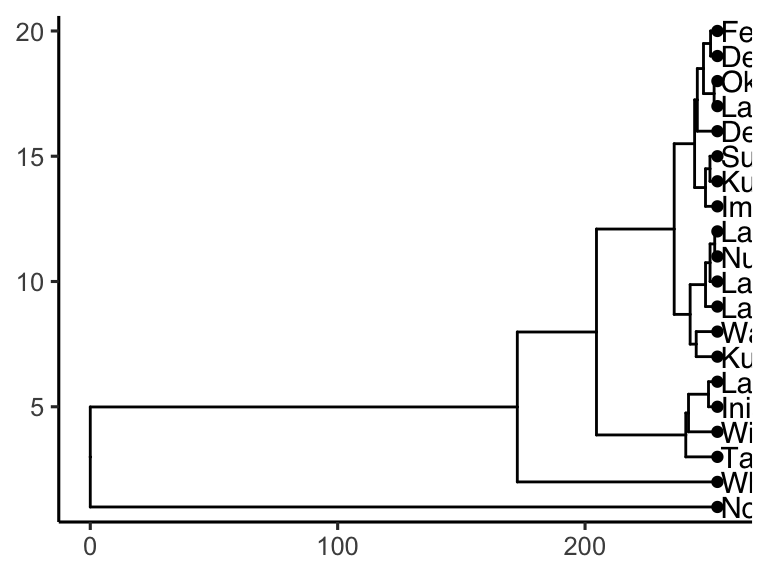
Very nice!
further reading
For more information on plotting annotated trees, see: https://yulab-smu.top/treedata-book/chapter10.html.
For more on clustering, see: https://ryanwingate.com/intro-to-machine-learning/unsupervised/hierarchical-and-density-based-clustering/.
exercises
For this set of exercises, please use runMatrixAnalysis() to run and visualize a hierarchical cluster analysis with each of the main datasets that we have worked with so far, except for NY_trees. This means: algae_data (which algae strains are most similar to each other?), alaska_lake_data (which lakes are most similar to each other?). and solvents (which solvents are most similar to each other?). It also means you should use the periodic table (which elements are most similar to each other?), though please don’t use the whole periodic table, rather, use periodic_table_subset. Please also conduct a heirarchical clustering analysis for a dataset of your own choice that is not provided by the source() code. For each of these, create (i) a tree diagram that shows how the “samples” in each data set are related to each other based on the numerical data associated with them, (ii) a caption for each diagram, and (iii) describe, in two or so sentences, an interesting trend you see in the diagram. You can ignore columns that contain categorical data, or you can list those columns as “additional_analyte_info”.
For this assignment, you may again find the colnames() function and square bracket-subsetting useful. It will list all or a subset of the column names in a dataset for you. For example:
colnames(solvents)
## [1] "solvent" "formula"
## [3] "boiling_point" "melting_point"
## [5] "density" "miscible_with_water"
## [7] "solubility_in_water" "relative_polarity"
## [9] "vapor_pressure" "CAS_number"
## [11] "formula_weight" "refractive_index"
## [13] "specific_gravity" "category"
colnames(solvents)[1:3]
## [1] "solvent" "formula" "boiling_point"